A team of scientists from Mount Sinai was able to uncover specific tumor microenvironment (TME) regulators using a novel functional genomics technology known as Perturb-map.
A team of scientists from Mount Sinai was able to uncover specific tumor microenvironment (TME) regulators using a novel functional genomics technology known as Perturb-map. The novel relationships discovered could have significant implications for cancer research.
Their research was recently published by Cell. “Our focus for this project was related to immunology as well as gene therapy,” Dr. Brian D. Brown, lead author and director of the Ichan Genomics Institute at Mount Sinai, told BioSpace. “Cancer immunology and cancer immunotherapy allow us to treat patients. Some patients have extremely effective responses to these therapies, but there are also many patients who don’t respond to them.”
Brown originally came into biomedical research with an interest in gene therapy. “I’ve always been interested in science fiction. I was excited about the idea of going into gene therapy, and it prompted me to get my Ph.D. The idea of not just treating disease, but curing it at a genetic level, was very exciting,” he said. “Right now, there’s a huge resurgence of interest in the gene therapy field. COVID vaccines are a result of that progress.”
Brown and his team have used CRISPR technology in their lab for years. “CRISPR can be powerful in gene therapy. One way it’s commonly used is to knock out genes. A number of years ago, scientists came up with ways to use CRISPR libraries, which include thousands of CRISPRs, to knock out specific genes and figure out what happens when each gene is knocked out. This tells us about the gene’s function,” he said. “What makes it so powerful is that you can knock out hundreds of thousands of genes simultaneously.”
He explained that while the CRISPR screens are extremely effective for cell-intrinsic research, they don’t give much information about a gene’s function outside of the cell. “There is an entire class of genes that function only outside the cell. CRISPR screens can’t assay for these types of gene functions. The way the screens are read out causes a loss of spatial resolution and a loss of information about what was around the cell.”
The researchers looked into the mechanism of tumors that do not traditionally respond well to treatment. “Tumors have a certain spatial arrangement of immune cells. Some of these cells are outside, while others are inside the tumor,” Brown said.
The structured arrangement of growing cancer cells, including the arrangement of B cells and T cells, has a large effect on patient outcome. “No matter what you do, T cells can’t kill cancer cells if they can’t get into the tumor. Patients with this kind of arrangement respond badly to treatment.”
In 2018, the lab invented a technology called Pro-Codes, or Protein Barcodes. “The technology is a way to barcode CRISPR libraries without having to read them with that next-generation sequencing. The beautiful thing is that this allows us to barcode cells with different CRISPRs and different gene knockouts. You get single-cell resolution,” Brown explained.
The Pro-Code system is the basis for their spatial functional genomics technology called Perturb-map. The team applied the Perturb-map platform to knock out genes in parallel in a mouse model of lung cancer.
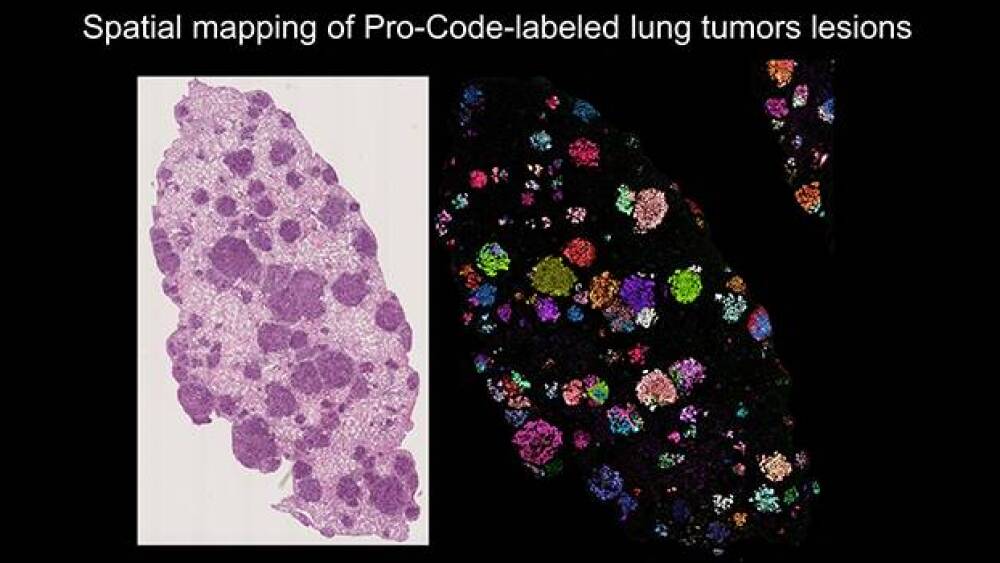
The scientists removed lung cancer cells from the mice, then analyzed stained tissue. They were able to identify the exact Protein Barcodes within the cells based on color staining alone. “The tumors were everywhere, but when we stained them, we could see each lesion lighting up with a different Pro-Code. We could see how homogeneous each lesion was for a particular barcode.”
The staining also allowed the scientists to differentiate between single-cell and multicell cloning. “We wouldn’t have known before that each tumor was started by one single cell, but we could see it here based on coloring. With our stained map, we could trace the cancer clones.”
In the past, the genes could be studied individually, but this technology allows researchers to study the effect of potentially thousands of genes on cancer at the same time. “The important thing is that we built a library of Pro-Code CRISPR vectors to target these genes,” Brown said. “These genes were indicated in tumor immunology already, but we haven’t been able to assay in this manner before. Each cell has an individual knockout, but this allowed us to study them in parallel. We could look at speed and understand how two knockouts would interact with each other. This is important in cancer immunology.”
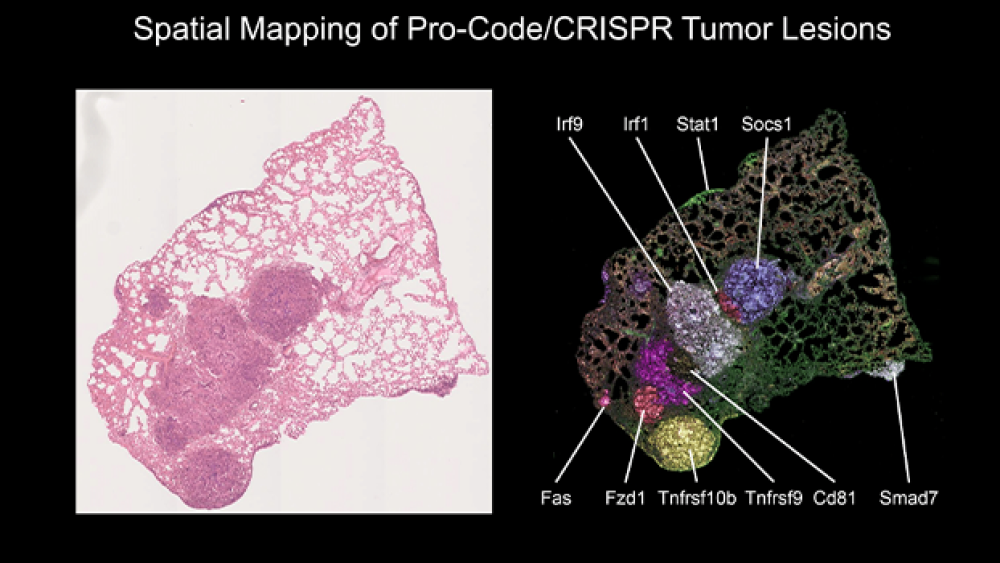
This image on the left shows a close-up of lesions in lung tissue before staining, and the image on the right shows the lesions after staining. “In the left picture, the lung tissue appears to have only one large tumor or a couple. But once we stained it, we could ask interesting new questions that we haven’t been able to ask before in functional genomic approaches. Before, we couldn’t even guess the exact number of tumors, and we couldn’t separate lesions. This image shows us the exact number,” Brown explained.
The researchers compared lung lobes from a number of mice to ascertain patterns. “You can determine the number of distinct lesions, their size and how they grow. You can determine if there are more or less immune cells infiltrating. This opens doors to much that we couldn’t do before. It’s quite powerful, especially for the cancer immunology field.”
The imaging allowed the researchers to determine that tumors grew much better, more frequently and bigger with specific genes. In comparison to control tumors, they collected data on morphology of the tumors by assessing large numbers of gene effects all at once. “This couldn’t be done with the other CRISPR approach or any other functional genomics approach,” Brown said.
The scientists were also able to look at immune cell infiltration. “We were able to map where each knockout was, and we could stain different immune cells. For example, we learned that a Socs1 knockout had more T cells than others. We learned that Tgfbr2 tumors grow the biggest.”
Being able to view the tumors side by side using the Pro-Code technology allowed the researchers to determine how localized a gene knockout’s effect could be on the TME. Brown explained, “These important findings can represent a pocket of resistance to immunotherapy. When we treat tumors with a drug, we see certain cells kill others, but these cells can’t get into the Tgfbr2 tumor. The T cells are being excluded from the cold tumor. Socs1 is considered a hot tumor. Hot tumors are more sensitive to chemotherapy. When we use screens with spatial recognition, we can see how a knockout influences response to a drug, and how knocking out a gene affects the local distal environment and location of a tumor.”
Brown believes the biological findings and technology can have many applications in target discovery. “We need drugs that can remodel the tumor microenvironment. This is a huge need. There are one or two really great drugs that can turn on T cells, but that’s not useful if the cancer is using an alternative mechanism. We’re trying to find out what these mechanisms are.”
He explained that the technology could be used in TMEs where the T cells are unable to get in. “It could be used to find out what genes are driving the immunosuppressive environment. Then we could allow the T cells in and allow them to kill the cancer cells. Beyond that, we want to find the genes that are mediating drug resistance. We can use these types of screens to treat animals with a drug, then see what tumors grow better or worse in relation to a knocked-out gene.”
The technology may be able to determine which patients are good candidates for immunotherapy. According to Johns Hopkins, only 15 to 20% of patients respond well to immunotherapy.
“There are a huge number of patients being treated who, for reasons that we don’t understand, are not responding to immunotherapy,” Brown noted. “We’d like to not have to give those patients those drugs for many reasons. They’re expensive and time-consuming. We’d like to put them on a therapy that will work better. Identifying the next class of drugs is important. With better predictors, doctors can more easily determine who should be put on immunotherapy, and who should not.”
Brown reminds that before eventual therapeutic approaches can be established, understanding the novel biology of these processes should be prioritized. “The new frontier right now in experimental biology and biosciences, especially in cancer, is spatial. We’ve introduced spatial functional genomics here. Everyone is so excited because this helps us to learn how different cells are influencing each other in a complex tissue environment. This is very different than in a petri dish and different than a single cell model. Even with the most advanced technology before, we were losing information.”
The Perturb-map approach allows for interaction-specific data. “The data showing how different knockouts are influencing the microenvironment and how clones may or may not influence each other is really interesting,” he said. “As a tumor grows, there are new mutations that affect the microenvironment. We can even see the differences between parent tumors and new regions. The ability to knock out particular genes and potentially disrupt negative mutations is interesting and important. We hope that the main takeaway from our work will be these important biological findings.”