NanoString Technologies, Inc., a leading provider of life science tools for discovery and translational research, announced that a peer-reviewed publication using the GeoMx® Digital Spatial Profiler is to be published in the journal Nature.
SEATTLE--(BUSINESS WIRE)-- NanoString Technologies, Inc. (NASDAQ: NSTG), a leading provider of life science tools for discovery and translational research, today announced that a peer-reviewed publication using the GeoMx® Digital Spatial Profiler (DSP) is to be published in the journal Nature. This paper describes using the GeoMx DSP to develop a foundational dataset to better understand the biological impact of SARS-CoV-2 (COVD-19) infection in human lungs, which may lead to the identification of new therapeutic interventions and prevention strategies.
This press release features multimedia. View the full release here: https://www.businesswire.com/news/home/20210504005265/en/
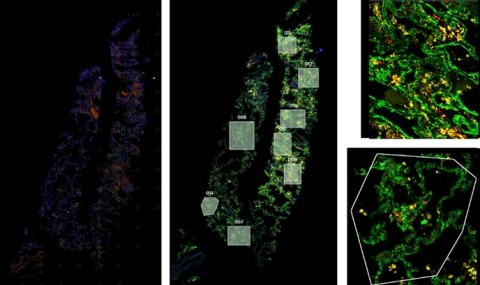
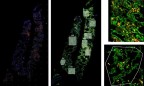
RNAScope (left) was utilized to assess the SARS-CoV-2 viral load of autopsy samples, and inform a strategy for region of interest selection (middle). Regions showing noteworthy biology were selected and segmented on CD45 and CD68 to focus on specific structures within the tissues (right). (Graphic: Business Wire)
Researchers from a network of institutions, including the Broad Institute and Beth Israel Deaconess Medical Center (BIDMC), Massachusetts General Hospital (MGH), the Ragon Institute of MGH, Massachusetts Institute of Technology (MIT), Harvard, Beth Israel Deaconess Medical Center, Brigham and Women’s Hospital, Columbia University Irving Medical Center, and scientists at NanoString used the GeoMx DSP to characterize autopsy lung tissue samples from patients who had been infected by the SARS-CoV-2 virus. In conjunction with single-cell RNA sequencing data collected for the project, researchers utilized the Cancer Transcriptome Atlas (CTA) and the Whole Transcriptome Atlas (WTA), with additional custom content specific for SARS-CoV-2 genes, to produce and spatially map a comprehensive cell-type atlas. The ability to pinpoint cellular processes, expression pathways, and immune cell profiles in this way offers key insights into systemic COVID-19 disease progression.
The study, to be published in Nature and led by Aviv Regev, Ph.D., a computational biologist and executive vice president of Genentech Research and Early Development, and professor at the Broad Institute of MIT and Harvard and the Department of Biology of MIT, is entitled “COVID-19 tissue atlases reveal SARS-COV-2 pathology and cellular targets.” [Broad Institute news release]
“Spatial profiling enabled us to expand our single cell studies and capture cellular profiles or transcriptional programs directly in the patient tissue,” said Ioannis Vlachos, Ph.D., Co-Director of the Bioinformatics Program, Cancer Research Institute, BIDMC, and co-senior author of the study. “The ability to capture more than 18,000 genes in situ using the “Whole Transcriptome Atlas” was truly transformative. We were able to understand how highly localized phenomena, inflammation, and the presence of SARS-CoV-2 virus can affect the cellular and transcriptional landscape of the COVID-19 lung, providing significant insights into the pathogenesis of COVID-19.”
“This research demonstrates the value of the GeoMx Digital Spatial Profiler in characterizing the tissue damage induced by SARS-CoV-2 infection,” said Dr. Sarah Warren, Senior Director of Translational Science at NanoString. “By working in partnership with the Broad Institute, Beth Israel Deaconess Medical Center, and other leading research centers, we have gathered meaningful information from the scientific community, creating a dataset of the impact of SARS CoV2 infection in multiple donors and the data will be made publicly accessible to accelerate scientific discovery.”
The GeoMx DSP enables researchers to rapidly and quantitatively characterize tissue morphology with a high-throughput, high-plex RNA and protein profiling system that preserves precious samples for future analyses. NanoString and its collaborators have presented DSP data in dozens of abstracts at major scientific meetings. This publication marks the milestone of being the 50th peer-reviewed publication, demonstrating DSP’s utility to address a wide range of biological questions in FFPE and frozen tissues. Interested parties can learn more about DSP by visiting https://www.nanostring.com/scientific-content/technology-overview/digital-spatial-profiling-technology.
NanoString launched its Technology Access Program (TAP) for the newly announced single and subcellular Spatial Molecular Imager to complement the existing TAP program for GeoMx. Under the program, a customer can submit tissue samples to NanoString to be analyzed using both spatial profiling platforms and receive a complete data package. Researchers interested in participating in NanoString’s Technology Access Program should contact the company at TAP@nanostring.com.
About NanoString Technologies, Inc.
NanoString Technologies is a leading provider of life science tools for discovery and translational research. The company’s nCounter® Analysis System is used in life sciences research and has been cited in more than 4,000 peer-reviewed publications. The nCounter Analysis System offers a cost-effective way to easily profile the expression of hundreds of genes, proteins, miRNAs, or copy number variations, simultaneously with high sensitivity and precision, facilitating a wide variety of basic research and translational medicine applications, including biomarker discovery and validation. The company’s GeoMx® Digital Spatial Profiler enables highly multiplexed spatial profiling of RNA and protein targets in a variety of sample types, including FFPE tissue sections. For more information, please visit www.nanostring.com.
NanoString, NanoString Technologies, the NanoString logo, GeoMx, and nCounter are trademarks or registered trademarks of NanoString Technologies, Inc. in various jurisdictions.
View source version on businesswire.com: https://www.businesswire.com/news/home/20210504005265/en/
Doug Farrell, NanoString
Vice President, Investor Relations & Corporate Communications
dfarrell@nanostring.com
Phone: 206-602-1768
Source: NanoString Technologies, Inc.
RNAScope (left) was utilized to assess the SARS-CoV-2 viral load of autopsy samples, and inform a strategy for region of interest selection (middle). Regions showing noteworthy biology were selected and segmented on CD45 and CD68 to focus on specific structures within the tissues (right). (Graphic: Business Wire)
View this news release and multimedia online at:
http://www.businesswire.com/news/home/20210504005265/en