The OpenFold group, a non-profit artificial intelligence research consortium of biotech and tech firms whose goal is to develop free and open-source software tools for biology and drug discovery, is announcing the funding of new large-scale protein studies at Prof. Gabriel Rocklin’s laboratory at Northwestern University.
New first-in-class datasets will improve the capabilities of state of the art protein AI models to create new biologic drugs
DAVIS, Calif.--(BUSINESS WIRE)-- The OpenFold group, a non-profit artificial intelligence (AI) research consortium of biotech and tech firms whose goal is to develop free and open-source software tools for biology and drug discovery, is announcing the funding of new large-scale protein studies at Prof. Gabriel Rocklin’s laboratory at Northwestern University. OpenFold is a project of the Open Molecular Software Foundation (OMSF), a non-profit organization advancing molecular sciences by building communities for open-source research software development. Prof. Rocklin’s lab is a pioneer in the creation of high-quality, large-scale, open protein data to improve AI models.
This press release features multimedia. View the full release here: https://www.businesswire.com/news/home/20231002433534/en/
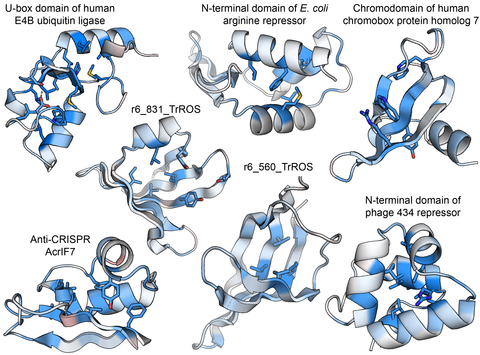
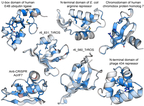
Figure Legend. Determinants of stability in natural and designed small protein domains. The TrROS domains are human designed, others are natural protein domains. Darker clue colors indicate that mutations are more destabilizing, red colors are more stabilizing. Figure from Tsuboyama et al. Nature 2023.
OpenFold released its first protein structure prediction models out of Prof. Mohammed AlQuraishi’s laboratory at Columbia University in Q2 2022, with speed and memory efficiency surpassing DeepMind’s earlier AlphaFold2, as well as the first public release of critical training code for protein structure prediction transformer models. These protein structure AI models are incredibly powerful for protein structure prediction, but have been found to have poor performance at predicting the influence of mutations on a protein’s stability and function. OpenFold and AlphaFold depend on the Protein Data Bank resource for learning to predict protein structures, but no similar resources currently exist for learning the principles of protein stability and function. Leveraging the power of deep learning in these areas will require new, innovative experiments that can generate the biophysical and functional data at the scale required to meaningfully train AI models.
Prof. Rocklin is a leader in the large scale analysis of protein function, stability and folding. Earlier in 2023, his lab introduced a powerful new method for measuring protein folding stability in the Nature article “Mega-scale experimental analysis of protein folding stability in biology and design” (Tsuboyama et al. Nature 2023). This work included folding stability measurements for nearly a million protein mutants, now openly released to the community. Researchers at over 50 universities and companies are already exploring these open data independently, and four new models have already been released that build on these data (For example, Dieckhaus et al. bioRxiv 2023.07.27.550881). This was an important first step toward understanding stability, but limitations in these data still make it challenging to develop fully general models.
In the new project funded by OpenFold, Prof. Rocklin will improve and expand these foundational studies. These new datasets will provide a never before seen level of detail on protein stability, enabling the training of new protein stability AI models with unprecedented accuracy, and much improved utility in protein design of novel biologic therapeutics.
“Prof. Rocklin’s experience combined with OpenFold’s state-of-the-art open source algorithms will set OpenFold’s first experimental collaboration up for success! We have seen that adding additional sequences to the models does not necessarily yield more accurate predictions, and we realize that real-world data is important to develop the next generation of more accurate AI models,” said Christina Taylor, Ph.D., Bayer Crop Science, Computational Molecular Design Lead and Science Fellow.
“Our lab is thrilled to work with OpenFold!” Professor Rocklin said. “Open data is a foundational resource powering the AI revolution in protein science, and we are completely aligned with OpenFold’s commitment to sharing and collaboration.”
About OpenFold
OpenFold is a non-profit artificial intelligence (AI) research consortium of academic and industry partners whose goal is to develop free and open-source software tools for biology and drug discovery, hosted as a project of the Open Molecular Software Foundation. For more information please visit: OpenFold Consortium
View source version on businesswire.com: https://www.businesswire.com/news/home/20231002433534/en/
Contacts
Press and membership inquiries should be directed to info@openfold.io
Source: Open Molecular Software Foundation
Figure Legend. Determinants of stability in natural and designed small protein domains. The TrROS domains are human designed, others are natural protein domains. Darker clue colors indicate that mutations are more destabilizing, red colors are more stabilizing. Figure from Tsuboyama et al. Nature 2023.
View this news release and multimedia online at:
http://www.businesswire.com/news/home/20231002433534/en